
Reads were down-sampled to 74000 reads with seqtk and aligned to SARS-CoV-2 reference genome (NCBI, NC_045512) with minimap2. Libraries were constructed using the NEBNext ARTIC SARS-CoV-2 Companion Kit (ONT) and sequenced on a GridION.
OXFORD NANOPORE MIDNIGHT PROTOCOL PLUS
Amplicons were generated from the same Omicron variant SARS-CoV-2 viral gRNA clinical sample using NEBNext ® VarSkip Short SARS-CoV-2 primer pools, NEBNext VarSkip Short v2 SARS-CoV-2 primer pools, MilliporeSigma ARTICv4, MilliporeSigma ARTICv4 primer pools plus IDT ARTICv4.1 spike-in primers (ARTICv4.1). Coverage depth per base was determined, reads were down-sampled to 0.5M PE reads with seqtk and aligned to SARS-CoV-2 reference genome (NCBI, NC_045512) with Bowtie2.įigure 4: Coverage of a SARS-CoV-2 Omicron clinical sample with updated primer schemes and Oxford Nanopore Technologies sequencingĬaption: Integrative Genome Viewer visualization of read coverage across the SARS-CoV-2 genome (log scale). Libraries were constructed using the NEBNext ARTIC SARS-CoV-2 FS Library Prep Kit (Illumina ®) and sequenced on a MiSeq ® instrument (2x75 bp).

The same Omicron variant SARS-CoV-2 viral gRNA clinical sample served as a template for all primer schemes except ARTICv3. Amplicons were generated using NEBNext ® VarSkip Short SARS-CoV-2 v2 primer pools, NEBNext VarSkip Short SARS-CoV-2 primer pools, NEBNext ARTIC SARS-CoV-2 primer pools (ARTICv3), MilliporeSigma ARTICv4 primer pools plus IDT ARTICv4.1 spike-in primers (ARTICv4.1), or MilliporeSigma ARTICv4 primer pools. Integrative Genome Viewer visualization of read coverage across the SARS-CoV-2 genome (log scale). The reagents for RT-PCR and library prep are optimized for the SARS-CoV-2 ARTIC workflow.įigure 3: Coverage of a SARS-CoV-2 Omicron clinical sample with updated primer schemes and Illumina sequencing These balanced primers provide greater uniformity of genome coverage (Figure 3) from 10-10,000 SARS-CoV-2 genome copies. NEBNext ARTIC kits include V3 ARTIC primer pools, which have been balanced using methodology developed at NEB based on empirical data from sequencing. NEBNext ARTIC SARS-CoV-2 Workflow for Oxford Nanopore Technologies Sequencing NEBNext ARTIC SARS-CoV-2 FS Library Prep Kit (Illumina) Workflow NEBNext ARTIC SARS-CoV-2 Library Prep Kit (Illumina) Workflow Express protocols are also available, with fewer clean ups.įor more detailed workflows, use the links below: In combination with optimized reagents for RT-PCR, the kits deliver improved uniformity of amplicon yields from gRNA across a wide copy number range.įigure 1: NEBNext ARTIC Standard Workflow Overview.

The two kits compatible with Illumina sequencing generate library inserts of ~150 bp or ~400 bp, for 2 x 75 or 2 x 250 sequencing, respectively (Figure 2). The NEBNext ARTIC SARS-CoV-2 RT-PCR Module contains only the reagents required for cDNA synthesis and targeted cDNA amplification from SARS-CoV-2 genomic RNA.Įxpress workflow options are provided, with reduced cleanup steps. The ARTIC method is a multiplexed amplicon-based whole-viral-genome sequencing approach (Figure 1), and the NEBNext ARTIC kit options are compatible with Illumina and Oxford Nanopore Technologies sequencing platforms. nCoV-2019 sequencing protocol v2 (GunIt)).
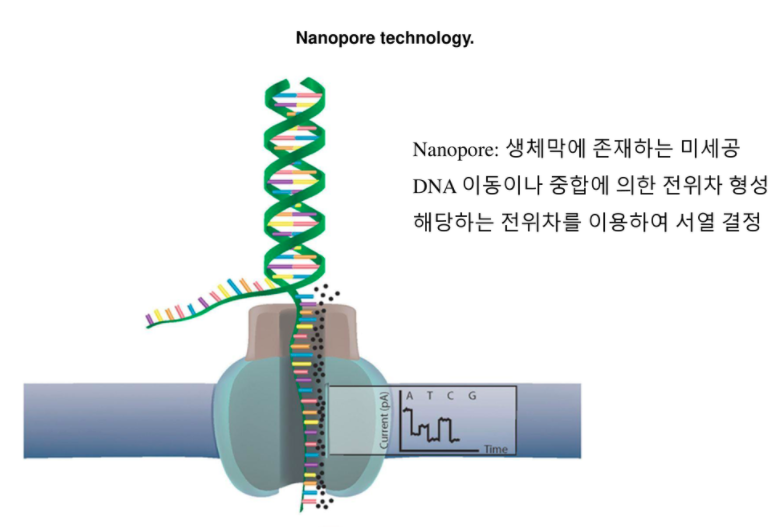
The NEBNext ARTIC kits are based on the original work of the ARTIC Network, who quickly adapted their protocols to SARS-CoV-2 (Josh Quick 2020. Especially with the ongoing emergence of SARS-CoV-2 variants that affect virus transmission and other metrics significant to public health, there is an increasing need for reliable, accurate and fast methods for sequencing SARS-CoV-2.


 0 kommentar(er)
0 kommentar(er)
